Well we looked up at the night sky briefly, just to notice that Venus and a little sliver of the Moon are very close together, but the main activity again this week is sheltering in place, where we looked more into the how the RT-PCR test for viruses works and some infection spread modeling terms.
First up if you have been following the YouTube lecture series on virology you probably have discovered the need for description of some of the molecular biology of cells and viruses as discussed in those lectures. For me, I had studied molecular biology years ago when I was intoxicated with the idea of going back to school and studying biophysics. Well, it turned out that I realized that I was not quite smart enough to do that graduate work, especially after I took an organic chemistry class and decided to drop out after a semester. But I still had a pretty good microbiology text that is still pretty useful for the beginning biology student, even though this 2008 edition is probably a bit out of date now!
 |
| This 1200+ page textbook is very good introduction to "Molecular Biology of the Cell" |
So, you can read about PCR or there are many freely available YouTube videos covering the topic. My take on these topics is not that of an expert, but best viewed as perhaps how a student might answer questions on a quiz. For more of the real details check out the actual references from which the screenshots below are abstracted.
So PCR is a technique to amplify a small quantity of DNA to very large quantities, sometimes like a billion times. It uses enzymes, called DNA polymerase, first discovered to occur naturally in bacteria, that can assemble DNA subunits to match an initial strand of DNA. So a sample to be analyzed is placed in a test tube and mixed up with DNA bases, polymerase, and a primer. The primer is a single stranded sample of DNA, of about 20-30 base pairs, known to be the expected sample. If the sample actually contains the primer somewhere in its much longer DNA sequence, then PCR will amplify the number of molecules present up into the billions.
 |
| Initial setup for conducting PCR analysis (Source: https://www.youtube.com/watch?v=DH7o9Df5_50) |
The first step of the PCR process is to heat the sample to break the hydrogen bonds so that the DNA strands separate into two separate strands. You can see that this step on takes about 30 seconds to cause the sample DNA to separate into two strands.
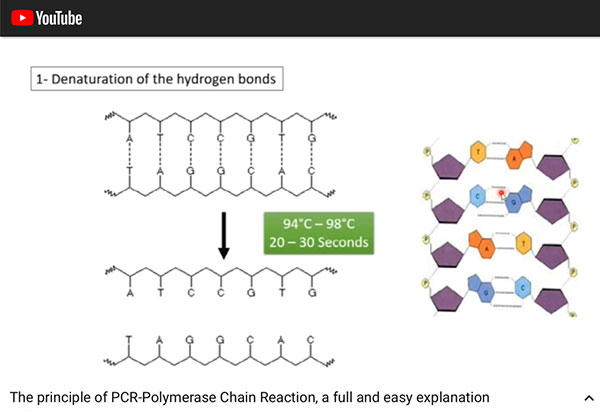 |
| Stage 1 of PCR process -- Denaturation of hydrogen bonds (Source: https://www.youtube.com/watch?v=DH7o9Df5_50) |
The next step involves cooling the sample down to about 50-65 degrees C so that the two primers can find and align up to the matching DNA sequence so that the sample and the primer can become hydrogen bonded. This step takes about 40 seconds. The selection of the primers allows one to choose the segment of DNA between the two primers to be the selected portion of the DNA sample that will be amplified. This is where you will see the effects of DNA replication starting from the 5' end to the 3' end takes place. I'm not discussing any of the 5' vs. 3' duplication of DNA, but you can read all about this amazing replication step in any of the referenced textbooks.
 |
| Step 2 of PCR process -- Annealing (Source: https://www.youtube.com/watch?v=DH7o9Df5_50) |
Next the temperature is increased a bit so that the elongation process can take place. This is when the other base pairs are added to the sample DNA. This is the first of may duplication steps, where once there was one strand of DNA and now there are two. Note that the two stranded DNA now is only that section of the sample DNA that was located between the two primer sections.
 |
| Step 3 of PCR process - Elongation (Source: https://www.youtube.com/watch?v=DH7o9Df5_50) |
So, that s the first of many thermal cycles. Each thermal cycle is now repeated automatically as many times to get the required amount of amplification.
 |
| Repeat the thermocycle many times to amplify the DNA (Source: https://www.youtube.com/watch?v=DH7o9Df5_50) |
So we see that at first there was one strand of DNA, then there are two, then four and so on until the end of the reaction. This slide shows that after 20 thermal cycles we will have amplified the initial sample of DNA by over a million times. All of this takes place in a little more than an hour.
 |
| Repeat thermal cycle for 20 times to get a million fold (Source: https://www.youtube.com/watch?v=DH7o9Df5_50) |
So, we started off with a small sample of DNA and have now multiplied it by a billion times or so. How do we analyzed the result? New techniques today probably rely more on fluorescent tags that are added to the sample as part of the PCR process and then you can just shine a laser beam through the amplified sample and get a very quick reading of the amount of DNA present. But, it used to be and perhaps still is used in much of biology research, that the amplified samples would be put on a slide with agar gel and then use electrophoresis to separate and identify the amount of DNA present. So how does this process work?
In this next screenshot by Joseph Ross we see how it is done. He uses an example where the sample uses DNA for kids and parents, but the main thing to keep in view is how the gel elecrophoresis separates the DNA by molecular weight. The samples are placed in wells in the gel and an electric potential is applied to the test strip. DNA is typically considered a more electrically negative molecule so that it is attracted more to the positive electrical terminal. So the DNA molecules move through the gel and get separated by molecular weight. The smaller DNA segments move faster through the gel. So at the end of the test you get a display of DNA by molecular weight. Several control paths are also built into the get strip. First is just a pure water sample and in this case there should be no visible bands of DNA because there is none in the pure water and if some bands show up then some contamination has occurred during the test. In addition a strip of four diluted amounts of standard DNA segments is also added so that you can tell what the molecular weight of any given band is. So here we see the resolution is among bands of 100, 250, 500 and 1000 DNA molecular weight units.
 |
| Gel electrophoresis analysis of DNA segments (Source: Joseph Ross, https://www.youtube.com/watch?v=PXxtJlLpcTM) |
So that is a brief story of how PCR is used to conduct testing of samples. Next lets look at a few key points that came up in the Virology course by Dr. Vincent Racaniello, Columbia U. I am still trying to make my way through this course, which has now got more complicated and is straining my limited background in microbiology. I am however, reminded again of the fascinating complexity that goes on in all living cells and of all of the chemical reactions that are necessary for life. Again, you can check out all of the course lectures for free on YouTube at:
https://www.youtube.com/playlist?list=PLGhmZX2NKiNldpyRUBBEzNoWL0Cso1jip&app=desktop
First we should recall what type of information is encoded in the viral genome. Remember that the current coronavirus contains RNA rather than DNA. Viruses are known to contain seven different types of encoding in their genome. Some use DNA and some use RNA. Those that use RNA must also include the coding for enzyme that will be released once the virus is in a living cell that will convert the viral RNA into a form of DNA that the infected cell can then, unfortunately, duplicate just using the same internal machinery used as a part of normal operations.
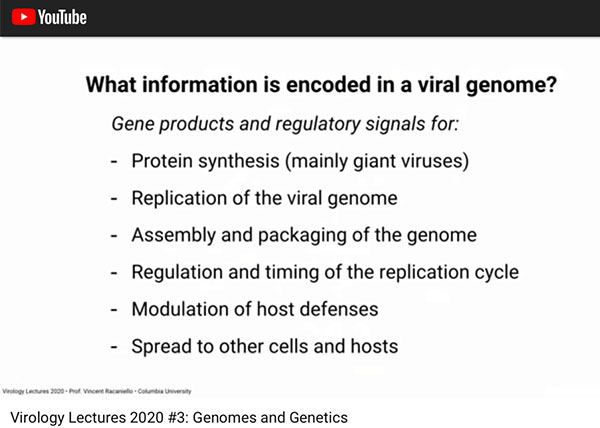 | |
Basic functionality (Source:
|
 |
| Portion of 29,903 base pairs of DNA sequence for current coronavirus (Source: www.ncbi.nih.gov) |
Also just in case you are wondering what the structure of the primers are that are used in the PCR process, you can see that in this screenshot from the WHO. For primers of length 20 bases, the number of possible orientations is 4 to the 20th power or about 1 trillion possible combinations. So, it is maybe possible for there to be more than just the one expected location in an RNA of 29,903 bases, but the chances are not great of selecting the wrong location.
 |
| Selection of primers used for PCR amplification of Coronavirus (Source: W. Bellini, www.WHO.int) |
Ok, so now back to the virology lectures where we should get some understanding of the how and why of social distancing requirements come about. Here you can see that it is the nature of water droplets expelled during a sneeze or cough and their size that determines when they will drop out of the surrounding air just do to gravitational settling. We are lucky that this happens. With other virus particles just breath alone can expel the particles over longer distances.
 | |
|
So understanding the size of the droplets and physical characteristics of the surround air can help predict the safe distance to prevent infection. In this laboratory study we see that temperature and humidity can also be factors. Remember it is not the case that if a virus lands on your skin that you will get infected. The virus must land on a live cell in order to infect the cell and our hands and other external skin are made of dead cells. So no infection, but we need to wash our hands so that we don't transfer the virus to our eyes or mouth or lungs where live cells are in direct contact.
 | |
|
We will often hear on the news statistics that talk about morbidity and mortality and other terms. What do all of these terms mean and how are they calculated? Well again the virology course defines all of these terms. Now as more data becomes available and more testing of the general population is completed we will get firmer numbers for all of these terms It is one thing to know the chances of getting infected, but it is a completely different thing to know the odds of having to go the hospital and once there the odds of the infection being fatal. Getting firmer numbers of all of these terms will enable us to set the path going forward.
 | |
|
Lastly, the term that is most important in stopping the epidemic is the basic reproductive number, R0. This number represents the number of cases that are potentially infected by one other person. In this screenshot you can see how R0 is defined and on what parameters it depends and the units in which is it is reported. If the R0 is greater than one, then each person infects at least other person and the epidemic can spread and increase. Our role is to help bring the number below one.
 |
| R0 definition and why we want it to be less than 1 (Source: Vincent Racaniello, Columbia, U.) |
Ok, enough of a student's version of virology and epidemiology. As we shelter in place, we have found the value of Zoom and other internet connectivity to enjoy a philosophy meetup, a physicist wannabe get together and even heard of a wine and cheese get together.
Until next time, let's do our part to drive R0 < 1,
Resident Astronomer George
Be sure to check out over 400 other blog posts on similar topics
If you are interested in things astronomical or in astrophysics and cosmology
Check out this blog at www.palmiaobservatory.com

No comments:
Post a Comment